How Do You Draw Tree for Hybridization
Abstract
Key message
Slash and burn practices affect tropical forests. Our results showed strong introgression between Inga ingoides and Inga edulis in the species contact area. Interspecific hybridization could be sought to improve yield and tolerance to flooding and further increase the economic potential of the poorly drained Amazonian soils and minimize deforestation.
Context
Inga species are important components of tropical American forests, as well as a local food source. Little is known about the genetic structure of these species; in particular the amount of introgression among species remains unknown.
Aims
We assessed the degree of genetic divergence and introgression among populations of I. ingoides (Rich.) Willd. and I. edulis Mart. (Fabaceae) from three Peruvian Amazon tributary rivers.
Methods
Using microsatellite markers we determined the genetic structure of populations using an analysis of molecular variance and a Bayesian analysis of population structure in areas affected by seasonal river fluctuations and in 'terra firme' forests.
Results
Overall genetic differentiation was weak. The degree of genetic variation was similar in the two species. A putatively strong introgression was detected between the two species and an intense gene flow was identified among populations. This indicates that an intense gene flow had happened in the past, leading also to a small differentiation among populations within species.
Conclusion
Selection of natural hybrids or artificial hybridization between I. edulis and I. ingoides could be applied to improve legume size and yield in the later species, while maintaining tolerance to flooding. Improved I. ingoides could be used in multipurpose agroforestry on open areas along the rivers, instead of using the usual slash and burn practice to create inland open areas.
Introduction
The Amazon drainage basin containing mainly lowland rainforest habitats is a major component of the Neotropical region, with more than 8 million km2 and about 25 million people (Junk and Piedade 2011). The riparian forests in the rain forest cover about 1 million km2, which corresponds to around 50 % of the basin's entire wetland area. The species-rich floodplain forests along the large Amazonian rivers are able to survive floods up to 10 m deep for as long as up to 8 months per year (Junk and Piedade 2011, and references therein). Increasing population density and human activity are destroying the forest landscape and inflicting a loss of biological diversity (Oliveira et al. 2007). Today, due to the continuing massive pressure exerted by farmers, cattle ranchers, and logging companies on the forests, new management concepts are urgently required to avoid the destruction of this unique forest type (Junk and Piedade 2011). The Peruvian Amazon tropical area (ca. 661,000 km2) suffered disturbance and deforestation at the average rate of 647 km2 per year from 1999 to 2005: 75 % within legally sanctioned areas, 64 % concentrated around the Ucayali logging centre Pucallpa, and 1–2 % occurred within natural protected areas (Oliveira et al. 2007).
The genus Inga Mill. (Fabaceae) comprises ca. 300 species of trees restricted to tropical America. Each region has preferred edible Inga species sold in large quantities in markets during the fruiting season (Pennington 1997). Inga edulis Mart., which occurs naturally on non-flooded or temporarily flooded sites, is a widely distributed and highly valued species in the Amazon region: it has been improved by human selection focusing on edible fruit, and cultivated as a fruit tree in Peru for millennia, and more recently in agroforestry systems (Pennington 1997). Inga ingoides (Rich.) Willd., a close relative of I. edulis, is used frequently in gardens and pastures for its edible fruit, and has ecological adaptability with potential use in a wide range of locations with limited conditions due to flood or poor soil drainage (Pennington 1997). Biodiversity conservation in the Peruvian Amazon along the riverside zones, while maintaining land user benefits, could be achieved by using this underutilized crop for food and fodder, avoiding slash and burn practices (Lander and Monro 2015). The neglected I. ingoides species could be considered as a multipurpose fruit tree species in agroforestry and other crop systems practiced in areas affected by periodical flooding. Production of fruit and timber from this species near rivers would be less costly, more sustainable and more forest-friendly due to: (1) easy accessibility for humans, (2) economy of transport, (3) nutrient input provided by periodical flooding, and (4) cultivation in forest buffer zones avoiding new forest sites colonization. Thus, the use of I. ingoides in open areas affected by periodical flooding could be achieved by genetic improvement through selection of natural hybrids or artificial hybridization with I. edulis and backcrossing, selecting for tolerance to flooding, legume size and yield, similar to the type of breeding achieved in the genus Eucalyptus (Potts and Dungey 2004). Interspecific hybrids of Eucalyptus have been used in forestry for decades, particularly in tropical and sub-tropical forestry, with plantations initially based on outstanding spontaneous hybrids. Selection was based on phenotype, followed afterwards by breeding programs based on manipulated hybrids (Potts and Dungey 2004). A similar approach, initiated with the selection of performing hybrids, could be applied to the Inga species under study.
Population genetic studies of tropical trees have shown that most of the species investigated are outcrossed and exhibit high levels of genetic diversity and gene flow, carrying much of the variation within, rather than among populations (Finkeldey and Hattemer 2007, and references therein). Also, the specific evolutionary history of each species has played an important role in determining the level and distribution of genetic diversity (Hamrick et al. 1992). In tropical forests, the levels of genetic diversity within populations vary considerably among species (Finkeldey and Hattemer 2007), from H e = 0.11 in Acer skutchii Rehd. (Mexico) (Lara-Gomez et al. 2005) to H e = 0.78 in Swietenia macrophylla King (Brazil) (Lemes et al. 2003), with both studies using microsatellites. Genetic differentiation among populations is slightly higher for tropical forest tree species than for temperate forests tree species, probably due to higher fragmentation levels in tropical trees. Moreover, tropical tree species with abiotic seed dispersal show, on average, much higher differentiation among populations than biotic-seed dispersed species. Seed dispersal by animals (zoochory) is usually very efficient and results in low genetic differentiation among populations (Loveless 1992). In the genus Inga, few genetic diversity studies have been reported to date. Studies in I. edulis and I. vera, using microsatellite markers, compared natural vs. planted populations to understand habitat fragmentation and to clarify the impact of species domestication and possible diversity loss (Cruz-Neto et al. 2014; Hollingsworth et al. 2005; Dawson et al. 2008). The authors of the latter study found that diversity was lower in planted compared to natural populations, but the values were still relatively high and the genetic diversity in planted stands can, to some extent, be restored by receiving pollen from natural populations. To the best of our knowledge, no studies about the genetic diversity in I. ingoides have been published.
The present study, using microsatellite markers, focused on two main objectives: firstly, we wanted to study the genetic structure of the populations of I. ingoides and I. edulis, and secondly, based on the obtained genetic structure, we wanted to infer the suitability of a hybridization program. The specific aims of the present study were: (1) to test if populations from three Peruvian Amazon tributary rivers, geographically separated, had diverged and accumulated substantial differentiation among populations within the I. edulis and I. ingoides species; (2) to compare the genetic diversity and divergence of three natural I. ingoides populations with those of nearby I. edulis natural populations; (3) to check for putative introgression between both species; and (4) to discuss the possibility of the targeted hybridization between the two studied species, the transfer of the tolerance to flooding from I. ingoides to I. edulis, and the transfer of legume size and yield potential from the latter to I. ingoides.
Material and methods
Plant material and study site
The two sympatric Inga species were identified according to morphological aspects detailed in the online resource ESM_1.pdf (Pennington 1997). Inga ingoides is distributed from the Lesser Antilles and tropical South America to Bolivia, including coastal Brazil to southern Minas Gerais. Inga edulis and I. ingoides are sympatric species with overlapping distribution, but the former is more likely to be found in non-flooded sites since it can withstand only temporary floods. According to Pennington (1997), I. ingoides flowering season, from August to November, partially overlaps the I. edulis June–October flowering season. The Inga species has brush-type flowers with mainly nocturnal anthesis specialized for hawkmoth (Sphingidae) and bat (Phyllostomidae) visits (Cruz-Neto et al. 2011, and references therein), yet diurnal visits by hummingbirds (Trochilidae) and hawkmoths were also observed by Koptur (1984).
Plant material from 77 I. ingoides and 62 I. edulis individuals used in this study was collected in riparian situations along three Amazon River tributaries and in upland forests (Table 1; Fig. 1a, b) from 2009 to 2012. The RPI and RPE populations (hereafter, the first two letters of the population name are the initials derived from the site name, the third letter means I =I. ingoides and E =I. edulis) were sampled from original vegetation along the river Pacaya. The RSI and RSE populations were observed in original vegetation on the upper Samiria river. Both rivers belong to the protected area called Pacaya Samiria National Reserve (Fig. 1a). The RUI and RUE populations were sampled on secondary vegetation along the Utiquinia river from the San José village, situated on non-inundating terraces, to the periodically flooded and poorly drained sites heading downstream to the Ucayali river. The MAE population was sampled in the Macuya Experimental Forest, a 'terra firme' forest remnant, protected by the National University of Ucayali, surrounded by deforested logged areas close to the town of Von Humboldt. The SDE population was observed behind the Contamana city's secondary vegetation, which begins in undulated terrain and continues to the original vegetation in the protected mountain range called Sierra del Divisor National Park.
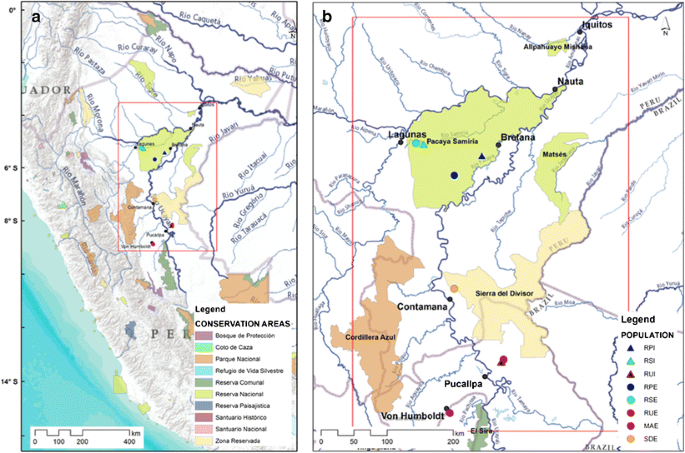
a Map of South America highlighting the study area. b Map with the rivers location, conservation areas and sampled populations located in the Samiria (RSI and RSE), Pacaya (RPI and RPE), and Utiquinia (RUI and RUE) rivers, and, also, the MAE and the SDE populations
The sampled trees were selected randomly and the minimum average distance between two sampled individuals from the same species was 200 m. Young leaves were collected from sexually mature trees and preserved in silica gel for further DNA extraction. Voucher specimens were archived in the Regional Herbarium of Ucayali IVITA-Pucallpa, Peru, with the code AR1-384.
DNA extraction and amplification
Total genomic DNA was extracted from dried young leaves with the Invitek, Invisorb® Spin Plant Mini Kit (http://www.stratec.com) according to the manufacture's instructions. We used four microsatellite primers, one (Pel5) primer was developed for Pithecellobium elegans Ducke by Daynandan et al. (1997), and the remaining three primer pairs (Inga03, Inga08 and Inga33) were developed by Hollingsworth et al. (2005) for I. edulis. A fluorescent dye (6-FAM, NED or VIC) was added to the 5′ end of each forward primer.
Loci were amplified individually in 10 μl reaction containing: 20 ng template DNA, 5 μM forward and reverse primer, 50 μM dNTPs, 2 mM MgCl2, 2 μl 5x GoTaq Flexi Buffer (Promega, Madison, WI) and 1.0 U GoTaq® Flexi DNA Polymerase (Promega). Amplifications were undertaken in Biometra® T1 Thermocycler (http://www.biometra.de/) using the following profile: 95 °C for 2 min; 95 °C for 15 s, 55 °C (Inga03) and 59 °C (Inga08, Inga33 and Pel5) for 30 s, 72 °C for 30 s, 30 cycles; 72 °C for 15 min. Completed reactions were loaded onto an ABI PRISM 310 Genetic Analyzer (Applied Biosystems, Foster City, CA) and run according to the manufacturer's protocol. Allele sizes were determined using the ROX500 internal size standard and the GeneMarker® v2.4 software (Applied Biosystems).
Data analysis
The diversity parameters comprised the number of alleles (N a), the effective number of alleles (N e), the observed heterozygosity (H o), the expected heterozygosity (H e) (Nei 1987), and the fixation index (F IS) (Weir and Cockerham 1984). A principal coordinate analysis (PCoA) was computed based on the pairwise Nei's genetic distance matrix. The analyses were performed using GenAlEx 6.5 (Peakall and Smouse 2012), except for the allelic richness (A R), which was computed using FSTAT 2.9.3 (Goudet 1995). Using the Genepop 4.3 software (Rousset 2008), the Hardy-Weinberg equilibrium (HWE) was tested for each population and locus (Markov-Chain method), the linkage disequilibria (LD) tests were done for all loci combinations, and the average frequency of null alleles were computed per population.
The grouping structure was further explored using a locus-by-locus analysis of molecular variance (AMOVA), implemented with the Arlequin 3.5 software (Excoffier and Lischer 2010). We estimated the variance components and genetic variation using a non-hierarchical and hierarchical analysis considering all of the populations or the two groups (species), respectively. The significance values were computed by a permutation test from 1,000 permuted matrices.
A Bayesian clustering method was carried out using the STRUCTURE version 2.3.3 software (Pritchard et al. 2000) to estimate the number of genetic clusters (K) and to fractionally assign individuals of both Inga species to the inferred groups. We applied the model which allows population admixture and correlated allele frequency. The K was set from one to eight, and the simulation was run ten times at each K value to confirm the repeatability of the results. Each run comprised a burn-in period of 25,000, followed by 100,000 Markov chain Monte Carlo (MCMC) steps. Afterwards, the STRUCTURE output data were parsed using the program Structure-sum (running under the R platform) (Ehrich et al. 2007), mainly to determine the optimal K value following Nordborg et al. (2005) and Evanno et al. (2005) methods. Therefore, we used the ΔK distribution statistic of Evanno et al. (2005) to determine the most appropriate number of genetic clusters through the detection of the second rate of change in LnP(D). In addition, the similarity coefficient between ten structure runs was computed, and for values higher than 0.9 we assumed that each run ended with a similar result. An alignment of cluster assignments across replicate analyses was then conducted in the CLUMPP 1.1.2 software (Jakobsson and Rosenberg 2007), and subsequently visualized using DISTRUCT 1.1 (Rosenberg 2004).
Results
Genetic diversity and inbreeding
The four simple sequence repeat (SSR) loci used in this study were very polymorphic, with a total of 66 alleles in I. ingoides and 58 alleles in I. edulis. However, the higher number of alleles (N a) could reflect the higher number of individuals (N) in some of the populations in both species: RPI (N = 47; N a = 13.3) and MAE populations (N = 27; N a = 11) (Table 2). The effective number of alleles (N e) was higher in the I. ingoides southern population, RUI (6.1), and lower in the northern one, RSI (4.4). The I. edulis western population (MAE) held the highest N e value (6), and the smallest value was found in the eastern SDE population (2.8) (Table 2).The rarefaction method displayed similar average allelic richness (A R) values in both species (5.1) (Table 2), due to differences in sample size per population.
The expected heterozygosity (H e) was also similar in both species (ca. 0.70), but the observed diversity (H o) was lower for I. ingoides (0.54) compared with I. edulis (0.68), which leads to a positive inbreeding coefficient (F IS) in the former (Table 2). All the I. edulis populations are in Hardy-Weinberg expectations (HWE), but not the I. ingoides populations (Table 2). High F IS values—the loss of heterozygosity due to non-random mating of parents—reflected differences between observed and expected heterozygosity. I. ingoides populations (RPI, RSI and RUI) departures from HWE showed significant (P < 0.001) heterozygote deficiency. On the contrary, the I. edulis populations F IS values were not significant. The average frequency of null alleles was similar and low in both species. In addition, no linkage disequilibrium was detected between different genotypes with the Fisher exact test among the different loci (P > 0.05), indicating that all four loci segregate independently of each other in both studied species.
The loci with higher Na (18) were different in both species: Pel5 in I. edulis, and Inga03 and Inga33 in I. ingoides (Table 3). The A R per loci ranged from 4.2 (Inga08) to 11.5 (Inga33) based on the minimum sample size of 14 individuals in I. ingoides, and from 3.3 (Inga08) to 7.14 (Pel5) based on the minimum sample size of 5 individuals in I. edulis (Table 3). The Inga08 locus had the lowest H e values in both species (0.24 and 0.47, in I. ingoides and I. edulis, respectively), and the Pel5 locus had the highest value (ca. 0.90).
Private alleles (P a) were identified for each I. ingoides population, the highest P a per population was found in the RPI population (3.5 across loci) and the lowest value in the RSI (0.75). The locus Inga03 had the highest P a (2.7 across all populations) and Inga33 had the lowest (1.33) in this species (ESM_2.pdf). P a were identified in four I. edulis populations and the RPE had the highest P a (1.25 across loci). The SDE population had no private allele, probably due to the low N. Only two alleles are common to the RPI/E pair, in the other pairs there are no common private alleles. The populations RUI and RSE hold the highest N/NP a ratio, i.e., they have the highest number of private alleles compared to the population size (ESM_2.pdf).
Population differentiation and Bayesian cluster analysis
The PCoA analysis reveals populations' weak grouping (Fig. 2), with the first and the second factor explaining 68 % and 15 % of the total variation, respectively. The AMOVA revealed an overall low among population variation (ΦST = 0.05: P < 0.0001), and the highest variation of the data set was found within populations (94 %) (Table 4). Undoubtedly, group (a), including all the I. edulis populations, clustered separately from group (b), the three I. ingoides populations (Fig. 2). Furthermore, the AMOVA confirmed a low, yet significant (P < 0.02) differentiation between the two Inga species ΦCT = 0.036 (Table 4). The I. ingoides populations at the three different rivers were clearly separated, as observed in Fig. 2, widely separated along the second axis, although only explaining a small part of the variation. Indeed, the variation among populations within species was weak, ΦSC = 0.027 (Table 4).
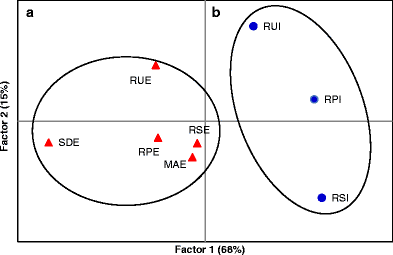
Principal coordinates analysis (PCoA) based on the Nei's pairwise genetic distances of I. edulis (filled triangles) and of I. ingoides populations (filled circles). Group a and group b, included populations from both species along the Pacaya, Samiria and Utiquinia rivers, respectively. The population SDE is an outlier
STRUCTURE distinguished clusters and the mean likelihood indicated two peaks at K = 2 and K = 4 (ESM_3A.docx). Additionally, we found that the mean similarity coefficient, the similarity between the ten runs, was consistently higher for K = 2 (ESM_3C.docx). Considering K = 2, the clusters corresponded to the two species groups, which had a biologically meaningful result: a clear introgression between species (Fig 3a).
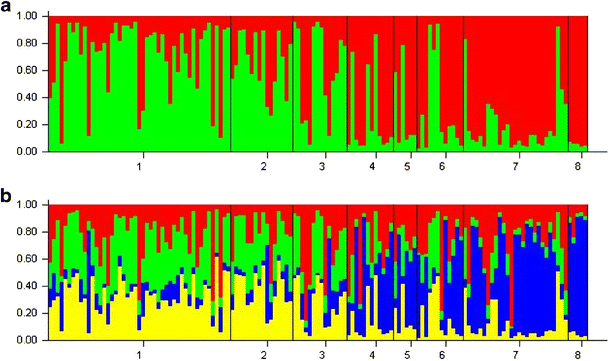
a Proportion of genotype membership q (y-axis) based on Bayesian cluster analysis. Each individual is represented by a single vertical line that is partitioned in different colors based on its genotype affinities to each cluster (K). Grey lines indicate the division between populations. Populations: 1 RPI, 2 RSI, 3 RUI, 4 RPE, 5 RSE, 6 RUE, 7 MAE, 8 SDE. b Plots of proportional group membership for the 139 trees for K = 2. Green Cluster 1, red cluster 2. b Plots of proportional group membership for the 139 trees for K = 4. Yellow Cluster 1, blue cluster 2, green Cluster 3, red cluster 4
Using the delta K criterion, the Bayesian clustering suggests the most probable presence of four groups (ESM_3B.docx), yet all individuals with mixed ancestry. Thus, the genetic clusters uncover extensive gene flow among populations. The mixed ancestry was particularly evident in the close population pairs along the rivers, with the more isolated I. edulis MAE and SDE populations clearly less mixed (Fig. 3a,b). The RUI/RUE populations seem to be the most mixed pair. The genetic clusters did not correspond closely to the morphological species, which suggest that gene flow has occurred between the species. The three I. ingoides populations seem to have the highest proportion of genotype affinities (or proportion of genotype membership) to both cluster 1 and 3, whereas I. edulis predominant proportion of genotype membership arises from cluster 2, in particular for the MAE and SDE populations (Fig. 3b). For K = 2, the mean introgression was higher for I. ingoides (25 %) than for I. edulis (18 %), considering the number of individuals with more than 50 % probability as belonging to the other species (q > 50 %); however the species introgression appears to be bidirectional (Fig. 3a). Nevertheless, if we consider only the populations along the rivers (RPE, RSE and RUE) the average introgression sums up to 28 % in I. edulis, and the MAE and SDE populations have negligible values. The RUI population has the highest introgression degree (36 %), almost twice the other I. ingoides populations (Fig. 3a).
Discussion
Genetic diversity
All populations displayed high values of expected heterozygosity (mean H e ~ 0.70, A R = 5.1). These estimates were slightly lower than estimates in natural populations of tropical trees I. vera (H e = 0.87; A R = 7.7) (Cruz-Neto et al. 2014), Symphonia globulifera L. (H e = 0.89) (Dick and Heuertz 2008) and Swietenia macrophylla King (H e = 0.78) (Lemes et al. 2003), but were very similar to the expected heterozygosity estimated for I. edulis by Hollingsworth et al. (2005) in the same region (Peruvian Amazon) (H e = 66 %). Normally, high levels of genetic diversity are maintained by high levels of gene flow facilitated by efficient pollen movement and the widespread occurrence of efficient self-incompatibility mechanisms (Dick et al. 2008). Some studies have demonstrated that some Inga species are obligate outcrossers, dependent on cross pollination to set fruits and seeds (Koptur 1984; Cruz-Neto et al. 2014) (see following section).
Inbreeding values differed in both species. Whereas I. edulis fits the low inbreeding values found in the I. vera natural populations' study using the same set of molecular markers (Cruz-Neto et al. 2014), our analyses revealed that the heterozygote frequencies in I. ingoides depart from the HWE, indicating either the existence of population substructure (due to the presence of genetically isolated groups, inbreeding, and/or spatial genetic structure) or null alleles. Since the estimated average frequency of null alleles is similar in both I. edulis and I. ingoides, we hypothesize that these differences could be explained by demography characteristics due to habitat preferences. The observed results may reflect I. ingoides's pioneer ability. This species rapidly colonizes the forest gaps opened by the seasonal river fluctuation, which results in populations being formed by patches of related individuals with a highly significant deficiency in heterozygotes due to recurrent biparental inbreeding. Thus, the heterozygotes deficiency could lead to lower competition ability, possibly explaining why this species is rarely found outside the riparian zone. In Acacia senegal (L.) Willd., Omondi et al. (2010) found that the only population with positive F IS was even-sized, suggesting the existence of one or few cohorts, possibly established together as a result of some disturbance event, and they argued that the area was prone to flooding, which could provide a mechanism for non-random seed dispersal. Indeed, seeds dispersed downstream could help to explain the departure from HWE in I. ingoides, though this hypothesis ought to be tested using a similar approach found in the study made with Calycophyllum spruceanum in the Peruvian Amazon (Russell et al. 1999).
The differences found in I. ingoides N e, a slightly higher value in the southern (RUI) population compared to the lower value in the northern population (RSI), may reflect altitudinal and flood pulse intensity differences, but may also reflect the high inbreeding value in RSI (whether the latter reason is the cause or the consequence will be difficult to disentangle). Indeed, I. ingoides tend to have a higher effective population size in less flooded southern areas than in those with higher river seasonal fluctuation, despite the species' tolerance to flooding, possibly due to lower biparental inbreeding. In the case of I. edulis, the highest N e value was found in the western MAE population, and the lowest in the eastern SDE population. The former population, situated closer to the Andean slopes, has a more favorable location than lesser elevated eastern sites prone to flooding, but a lower value in the latter population is probably due to differences in the number of sampled individuals.
The number of private alleles in I. ingoides across loci was almost twice as high as in I. edulis for a similar number of sampled individuals (N), which may indicate the presence of more intense gene flow in the latter species, in agreement with negligible inbreeding values. Within species, the number of private alleles seems to reflect N to a certain extent. Yet again, RUI has more than twice the P a than RSI, for comparable N; this might be the result of a higher inbreeding value due to putative higher parental inbreeding and consanguinity in the RSI population.
Genetic structure and putative species introgression
The partition of genetic variance in our studied species (94 % of the variance is observed within populations and a low genetic structure is detected among populations, 2.6 %), is very common in tropical forest tree species with high outcrossing rates, and among populations with high levels of gene flow (Finkeldey and Hattemer 2007). In a previous study, similar results were found with individuals showing mixed ancestry and low differentiation among populations, reflecting strong gene flow of Kenyan populations of Acacia senegal (Omondi et al. 2010). Within the genus Inga, Cruz-Neto et al. (2014) uncovered a similar pattern in the I. vera species.
Weak population genetic structure may be a consequence of the pollination system and also outcrossing in the populations under study. The majority of Inga species can be considered hawkmoth-pollinated, despite occasional visitation by bats and hummingbirds during the day (Cruz-Neto et al. 2014, and references therein). Hawkmoths, bats and hummingbirds can fly across large areas, ca. 15 km, during their foraging routes, carrying pollen grains to distant individuals (Koptur 1984). Pollen flow between distant individuals in different populations, due to pollinator behavior, contributed to high outcrossing rate and weak population substructure found in, e.g., I. vera natural populations (Cruz-Neto et al. 2014). Additionally, natural seed dispersal is performed by mammals and possibly birds that eat the sarcotesta and drop seeds elsewhere (Koptur 1984). Indeed, in a broad study with tropical tree species with abiotic seed dispersal (gravity dispersed and wind dispersed) showed, on average, much higher differentiation among population (G ST = 0.138) than animal dispersed species (G ST = 0.050) (Loveless 1992).
The weak population genetic structure together with the lack of isolation-by-distance (data not shown) suggests that species ecology, such as pollen and seed dispersal, and demographic history (impacted by flood) is a strong driver of population structure in the studied I. edulis and I. ingoides populations, as in the case of Acacia senegal (Omondi et al. 2010).
The Bayesian approach identified two to four clusters of genetically mixed individuals in both species, with higher admixture in those places where the two species were sympatric. Thus, we could assume that the populations were not reproductively isolated, and, probably, not well separated taxonomically. Nevertheless, some authors claim that some species of the Inga genus are cross-incompatible (e.g., Koptur 1984), but the data they presented does not support that conclusion, since the fruit set from hand cross-pollinated trees is clearly superior to the control.
Petit et al. (2004) reviewed the hybridization between two widespread and largely sympatric European oak species [Quercus petraea (Matt.) Liebl. and Q. robur L.]. They indicate that the parental taxa remain distinct, despite regular levels of gene flow between them, and emphasize the low differentiation found between both species. Yet, nuclear markers show more or less important differences in allelic frequencies between species. In another study, Moran et al. (2012) indicate that hybridization is pervasive in many plant taxa, with consequences for species taxonomy and local adaptation. They also indicate that oaks (Quercus spp.) are a paradigmatic case, since they are thought to hybridize readily yet retain distinct traits, drawing into question the biological species concept for such taxa, but the true extent of gene flow is controversial. Such reasoning could be extended to the Inga genus.
We should clarify that the morphological identification of all the individuals of the current study were rechecked with the key species identification clues according to morphology and no ambiguities were found. Selection against hybrids could hamper speciation in the Inga genus, but at least the past gene flow should be present in the individuals/populations in contact areas, which is the case of populations' species pairs: RUI/RUE, RPI/RPE and RSI/RSE, except in the more isolated I. edulis MAE and SDE populations. Introgression may be facilitated when species co-occur in areas where no intermediate habitats exist between the species ranges (Moran et al. 2012, and references therein). In our studied species, it seems that the opportunity for introgression should be close to the riverside, since I. edulis is relatively flood tolerant, and I. ingoides is probably more shade intolerant, or at least less competitive in this very harsh and competitive environment. Clearly the populations of I. edulis close to the rivers, where the two species overlap, suffer higher introgression, which is predictable due to the fact that the I. ingoides habitat is mainly found there. Endara and Jaramillo (2011) developed a study on the influence of microtopography on the distribution of Inga species. These authors indicate that one of the main factors explaining the distribution of the Inga species is the soil water content. Out the 16 more frequent Inga sympatric species they analyzed, 9 had a significant preference for one type of microtopography: "slope" and "ridge" (well drained) or "valley" (poorly drained soils). This fact indicates the importance of microhabitat to the sympatric species coexistence in the Inga species, and that edaphic specialization among species may create more available niches. Similarly, also in oaks, Q. robur appears to be more tolerant to soil anoxia than Q. petraea, and in mixed stands, succession towards the latter would be the rule, except under permanently humid conditions (Petit et al. 2004). Indeed, dynamic speciation through disruptive selection is also a hypothesis to be considered for the Inga species we studied.
In summary, we hypothesize that the opportunity for hybridization exists in the two Inga species studied here. Firstly, the natural distribution of the two species overlaps, although in our study the differences in habitat reflected the location of the sampled individuals of both species, with I. edulis found mainly in non-flooded terraces or temporarily flooded sites, and with I. ingoides found predominantly in periodically flooded areas (Pennington 1997). Secondly, in some studies based on I. ingoides and I. edulis, flowering phenology observations indicate synchronous flowering, which is also common in other Inga species (Pennington 1997; Cruz-Neto et al. 2011; Koptur 1984). Thirdly, the putative introgression between both species is also supported by low differentiation in microsatellite allele frequencies between the two co-occurring species (3.6 %), suggesting at least past gene flow (Moran et al. 2012). Lastly, both species are closely related from the genotypic point of view, which is also supported by the phylogenetic study done by Dexter et al. (2010), where they are found in the same node with 99 % support. In addition, speciation in the Inga genus is recent, and it is considered a classic example of a recent radiation with evidence for many species arising within the last 10 million years, some of them as recently as 2 million years ago (Richardson et al. 2001). Actually, due to a rapid and recent burst of diversification from the most recent common ancestor of the extant species, they found a poorly resolved phylogeny.
Suitability of a hybridization program
The use of wild hybrids and the establishment of a breeding program making use of the two species could bring important economical income to the periodically flooded arable lands in the Amazon basin with limited commercial use, with their potential incorporation into agroforestry systems. The ability of "pioneer" light-demanding species to grow in open spaces and inhospitable lands, could bring those species into the forefront of our concerns, by making flooded sites usable by flood-resistant and performing hybrids. Natural hybrids occur and are common in the species contact areas, according to our results, which are also indicative that artificial hybrids are possible in practice. Thus, natural hybrids' selection and/or artificial hybridization between I. edulis and I. ingoides could be applied to improve legume size and yield in the latter species, while maintaining tolerance to flooding. The success of the hybrids, and the development of these hybrids for commercial deployment, is dependent on two very important aspects. Firstly, hybrid variation and therefore selection within hybrids is dependent on the diversity of the parent species involved. Secondly, successful hybrid utilization is dependent largely on the vegetative propagation ability of the species (Potts and Dungey 2004). Our study revealed a high genetic diversity in both species, but care should be taken in avoiding related trees, particularly in the case of I. ingoides. We advise that future studies on hybridization and introgression in both species should be done together with flooding tolerance ability and legume and yield in hybrids testing, and wild hybrids could be procured by making use of today's available approaches, e.g., with tools developed specially for this genus by Dexter et al. (2010), which include both morphological and molecular approaches, and by Subashini et al. (2014) and Larcombe et al. (2014) in Eucalyptus. Also, vegetative propagation could be used to propagate hybrids, since Inga species can be propagated easily from semi-ripe branch cuttings, and, for example, I. edulis is considered an easy-to-root species (Pennington 1998).
References
-
Cruz-Neto O, Aguiar AV, Twyford AD, Neaves LE, Pennington RT, Lopes AV (2014) Genetic and ecological outcomes of Inga vera subsp affinis (Leguminosae) tree plantations in a fragmented tropical landscape. PLoS One 9. doi:10.1371/journal.pone.0099903
-
Cruz-Neto O, Machado I, Duarte J Jr, Lopes A (2011) Synchronous phenology of hawkmoths (Sphingidae) and Inga species (Fabaceae–Mimosoideae): implications for the restoration of the Atlantic forest of northeastern Brazil. Biodivers Conserv 20:751–765. doi:10.1007/s10531-010-9975-x
-
Dawson I, Hollingsworth P, Doyle J, Kresovich S, Weber J, Sotelo Montes C, Pennington T, Pennington R (2008) Origins and genetic conservation of tropical trees in agroforestry systems: a case study from the Peruvian Amazon. Conserv Genet 9:361–372. doi:10.1007/s10592-007-9348-5
-
Dayanandan S, Bawa K, Kesseli R (1997) Conservation of microsatellites among tropical trees (Leguminosae). Am J Botany 84:1658–1663
-
Dexter KG, Pennington TD, Cunningham CW (2010) Using DNA to assess errors in tropical tree identifications: How often are ecologists wrong and when does it matter? Ecol Monogr 80:267–286. doi:10.1890/09-0267.1
-
Dick C, Hardy O, Jones FA, Petit R (2008) Spatial scales of pollen and seed-mediated gene flow in tropical rain forest trees. Trop Plant Biol 1:20–33. doi:10.1007/s12042-007-9006-6
-
Dick CW, Heuertz M (2008) The complex biogeographic history of a widespread tropical tree species. Evolution 62:2760–2774. doi:10.1111/j.1558-5646.2008.00506.x
-
Ehrich D, Gaudeul M, Assefa A, Koch MA, Mummenhoff K, Nemomissa S, Consortium I, Brochmann C (2007) Genetic consequences of Pleistocene range shifts: contrast between the Arctic, the Alps and the East African mountains. Mol Ecol 16:2542–2559. doi:10.1111/j.1365-294X.2007.03299.x
-
Endara MJ, Jaramillo JL (2011) The influence of microtopography and soil properties on the distribution of the speciose genus of trees, Inga (Fabaceae:Mimosoidea), in Ecuadorian Amazonia. Biotropica 43:157–164. doi:10.1111/j.1744-7429.2010.00668.x
-
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620
-
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Eco Resour 10:564–567. doi:10.1111/j.1755-0998.2010.02847.x
-
Finkeldey R, Hattemer HH (2007) Tropical forest genetics. Springer-Verlag, Berlin, Heidelberg. doi:10.1007/978-3-540-37398-8
-
Goudet J (1995) FSTAT (Version 1.2): A computer program to calculate F-statistics. J Hered 86:485–486
-
Hamrick J, Godt M, Sherman-Broyles S (1992) Factors influencing levels of genetic diversity in woody plant species. New Forest 6:95–124
-
Hollingsworth PM, Dawson IK, Goodall-Copestake WP, Richardson JE, Weber JC, Sotelo Montes C, Pennington RT (2005) Do farmers reduce genetic diversity when they domesticate tropical trees? A case study from Amazonia. Mol Ecol 14:497–501
-
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806. doi:10.1093/bioinformatics/btm233
-
Junk W, Piedade MF (2011) An introduction to South American wetland forests: distribution, definitions and general characterization. In: Junk WJ, Piedade MTF, Wittmann F, Schöngart J, Parolin P (eds) Amazonian floodplain forests, vol 210, Ecophysiology, biodiversity and sustainable management. Ecological Studies. Springer, Dordrecht, pp 3–25. doi:10.1007/978-90-481-8725-6_1
-
Koptur S (1984) Outcrossing and pollinator limitation of fruit-set: Breeding systems of Neotropical Inga trees (Fabaceae: Mimosoideae). Evolution 38:1130–1143
-
Lander TA, Monro A (2015) Conservation of Brosimum alicastrum, an underutilized crop and keystone forest tree species; a potential win–win for conservation and development in Latin America. Biodivers Conserv:1–14. doi:10.1007/s10531-015-0913-9
-
Lara-Gomez G, Gailing O, Finkeldey R (2005) Genetic variation in isolated Mexican populations of the endemic maple Acer skutchii Rehd. Allg Forst Jagdztg 176:97–103
-
Larcombe MJ, Vaillancourt RE, Jones RC, Potts BM (2014) Assessing a Bayesian approach for detecting exotic hybrids between plantation and native Eucalypts. Int J Forest Res:13. doi:10.1155/2014/650202
-
Lemes MR, Gribel R, Proctor J, Grattapaglia D (2003) Population genetic structure of mahogany (Swietenia macrophylla King, Meliaceae) across the Brazilian Amazon, based on variation at microsatellite loci: implications for conservation. Mol Ecol 12:2875–2883
-
Loveless MD (1992) Isozyme variation in tropical trees: patterns of genetic organization. New Forest 6:67–94. doi:10.1007/bf00120640
-
Moran EV, Willis J, Clark JS (2012) Genetic evidence for hybridization in red oaks (Quercus sect. Lobatae, Fagaceae). Am J Botany 99:92–100. doi:10.3732/ajb.1100023
-
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
-
Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C, Zheng H, Bakker E, Calabrese P, Gladstone J, Goyal R, Jakobsson M, Kim S, Morozov Y, Padhukasahasram B, Plagnol V, Rosenberg NA, Shah C, Wall JD, Wang J, Zhao K, Kalbfleisch T, Schulz V, Kreitman M, Bergelson J (2005) The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol 3:e196. doi:10.1371/journal.pbio.0030196
-
Oliveira PJC, Asner GP, Knapp DE, Almeyda A, Galván-Gildemeister R, Keene S, Raybin RF, Smith RC (2007) Land-use allocation protects the Peruvian Amazon. Science 317:1233–1236. doi:10.1126/science.1146324
-
Omondi S, Kireger E, Dangasuk O, Chikamai B, Odee D, Cavers S, Khasa D (2010) Genetic diversity and population structure of Acacia senegal (L) Willd. in Kenya. Trop Plant Biol 3:59–70. doi:10.1007/s12042-009-9037-2
-
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539. doi:10.1093/bioinformatics/bts460
-
Pennington TD (1997) The Inga Genus Botany. Royal Botanic Gardens, London
-
Pennington TD (1998) Inga management. In: Pennington T, Fernandes E (eds) The genus Inga: utilization. Kew: Royal Botanic Gardens, London, pp 159–167
-
Petit RJ, Bodénès C, Ducousso A, Roussel G, Kremer A (2004) Hybridization as a mechanism of invasion in oaks. New Phytol 161:151–164. doi:10.1046/j.1469-8137.2003.00944.x
-
Potts BM, Dungey HS (2004) Interspecific hybridization of Eucalyptus: key issues for breeders and geneticists. New Forest 27:115–138. doi:10.1023/a:1025021324564
-
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
-
Richardson JE, Pennington RT, Pennington TD, Hollingsworth PM (2001) Rapid diversification of a species-rich genus of neotropical rain forest trees. Science 293:2242–2245. doi:10.1126/science.1061421
-
Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138. doi:10.1046/j.1471-8286.2003.00566.x
-
Rousset F (2008) Genepop'007: a complete re-implementation of the genepop software for Windows and Linux. Mol Eco Resour 8:103–106. doi:10.1111/j.1471-8286.2007.01931.x
-
Russell JR, Weber JC, Booth A, Powell W, Sotelo-Montes C, Dawson IK (1999) Genetic variation of Calycophyllum spruceanum in the Peruvian Amazon Basin, revealed by amplified fragment length polymorphism (AFLP) analysis. Mol Ecol 8:199–204. doi:10.1046/j.1365-294X.1999.00551.x
-
Subashini V, Shanmugapriya A, Yasodha R (2014) Hybrid purity assessment in Eucalyptus F1 hybrids using microsatellite markers. 3 Biotech 4:367–373. doi:10.1007/s13205-013-0161-1
-
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Acknowledgments
We thank the Servicio Nacional de Áreas Naturales Protegidas and José Navarro Pacaya-Samiria National Reservation Director for investigation authorization (N°004-2012-SERNANP-RNPS-J); National University of San Marcos, Veterinary Institute of Tropical and Highland Investigations, Regional Herbarium of Ucayali-Pucallpa, Peru and the Director Zoyla Mirella Clavo Peralta for processing and archiving the plant material. Thanks are also extended to N. Roque for help with Fig. 1 and to I. Salavessa for editing the English of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This study was funded by several research grants provided by the Bilateral Project 'Morphological and genetic diversity of indigenous tropical trees in the Amazon—model study of Inga edulis Mart. in Peruvian Amazon', Czech Academy of Sciences and CONCYTEC, Peru 2011–2012; The Internal Grant Agency of CULS Prague (No. 20145023, No. 20145005); The Scholarship National University of Ucayali, Peru. European Union Lifelong Learning Programme Erasmus Consortium—Practical Placement Scholarship (Certificate No. CZ-01-2009); The Foundation Nadace Nadání Josefa, Marie a Zdeňky Hlávkových, Czech Republic (Foundation Grant); and the Fundação para a Ciência e a Tecnologia grant UID/AGR/00239/2013 supported MMR.
Additional information
Contribution of the co-authors A.R. was in charge of the study; B.L. supervised the writing of the article, supervised Alexandr Rollo, and coordinated the project; D.H. participated in designing of the study and sample collection B.M. participated in the STUCTURE analysis, and in the interpretation of the results; J.A. Chia Wong participated in tree sampling, and helped in the species identification; C.S. supervised the genotyping, and participated partially in the genotyping; R.C. supervised and organized the genotyping; C. Q.-S. participated in the AMOVA analysis, and in the interpretation of the results; M.M.R. performed data analysis (genetic diversity estimates), the interpretation of the results, participated in the paper writing, and co-supervised A.R. All authors reviewed and commented on successive drafts of the paper.
Handling Editor: Ricardo Alia
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rollo, A., Lojka, B., Honys, D. et al. Genetic diversity and hybridization in the two species Inga ingoides and Inga edulis: potential applications for agroforestry in the Peruvian Amazon. Annals of Forest Science 73, 425–435 (2016). https://doi.org/10.1007/s13595-015-0535-0
-
Received:
-
Accepted:
-
Published:
-
Issue Date:
-
DOI : https://doi.org/10.1007/s13595-015-0535-0
Keywords
- Agroforestry
- Biodiversity conservation
- Introgression
- Inga
- Peruvian Amazon
- Microsatellites
Source: https://link.springer.com/article/10.1007/s13595-015-0535-0
0 Response to "How Do You Draw Tree for Hybridization"
Post a Comment